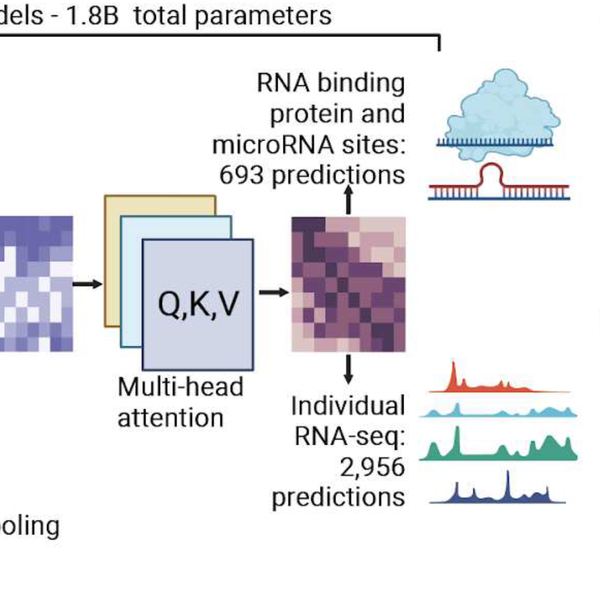
Deep Genomics Introduces the Most Advanced AI Foundation Model for RNA Disease Mechanisms and Candidate Therapeutics
Company’s unique AI foundation model BigRNA is the first transformer neural network for the discovery of RNA biology and therapeutics
Represents a new generation of deep learning AI that can be applied to a range of different RNA therapeutic discovery tasks
Comprised of nearly ...